Análise descritiva
## ------
## Welcome to AgroR!
##
## Shimizu, G.D.; Marubayashi, R.Y.P.; Goncalves, L.S.A. (2021). Package AgroR version 1.2.0
Um fator
data("pomegranate")
attach(pomegranate)
desc(trat,WL)
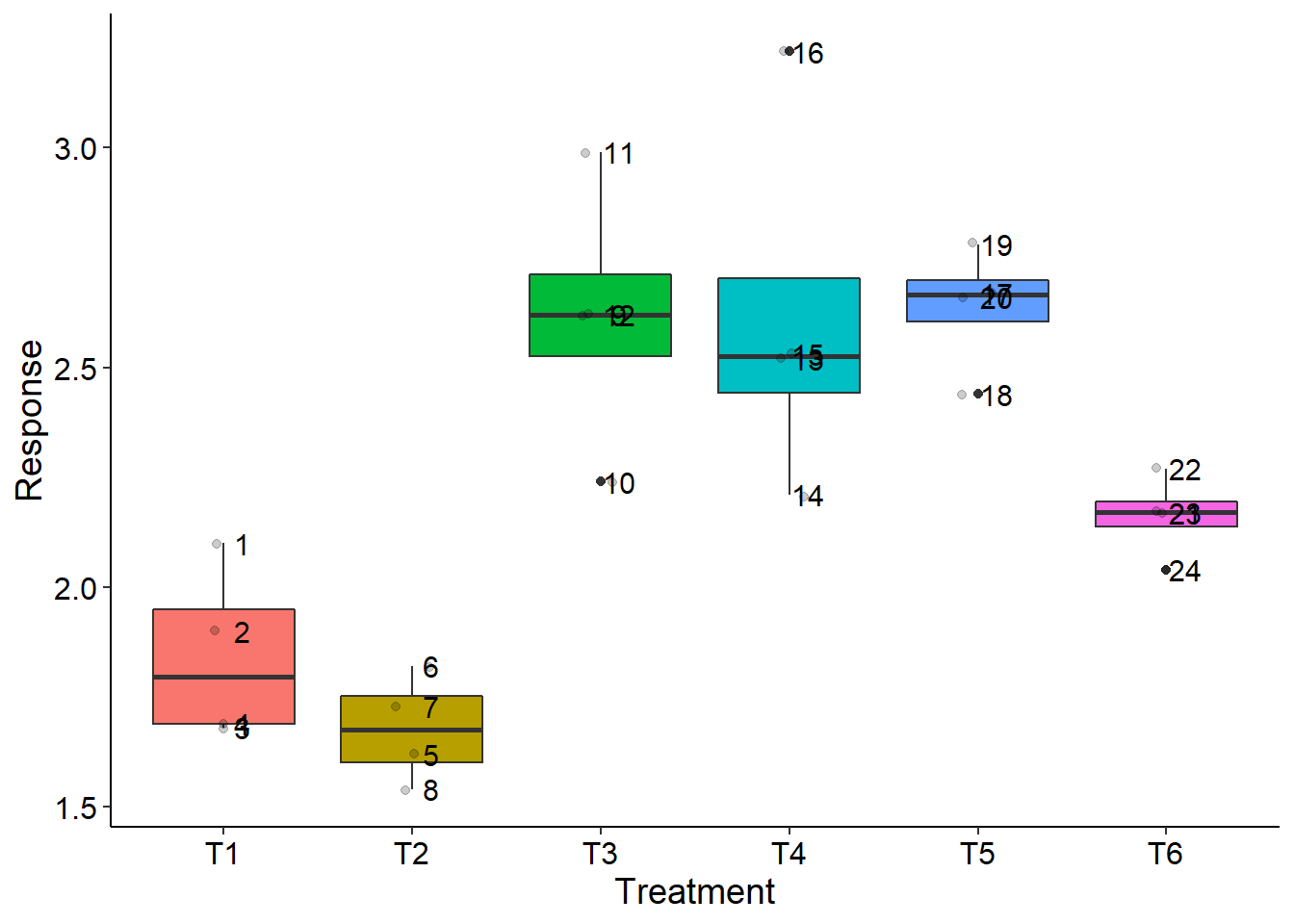
##
## -----------------------------------------------------------------
## General description
## -----------------------------------------------------------------
## Mean Median Min Max Variance SD CV(%)
## General 2.259583 2.225 1.54 3.22 0.207465 0.4554833 20.15784
##
## -----------------------------------------------------------------
## Treatment
## -----------------------------------------------------------------
## Mean Median Min Max Variance SD CV(%)
## T1 1.8425 1.795 1.68 2.10 0.039758333 0.19939492 10.821977
## T2 1.6775 1.675 1.54 1.82 0.015091667 0.12284814 7.323287
## T3 2.6175 2.620 2.24 2.99 0.093758333 0.30619983 11.698179
## T4 2.6200 2.525 2.21 3.22 0.182066667 0.42669271 16.285981
## T5 2.6375 2.665 2.44 2.78 0.020291667 0.14244882 5.400903
## T6 2.1625 2.170 2.04 2.27 0.008891667 0.09429563 4.360492
Dois fatores
data(cloro)
attach(cloro)
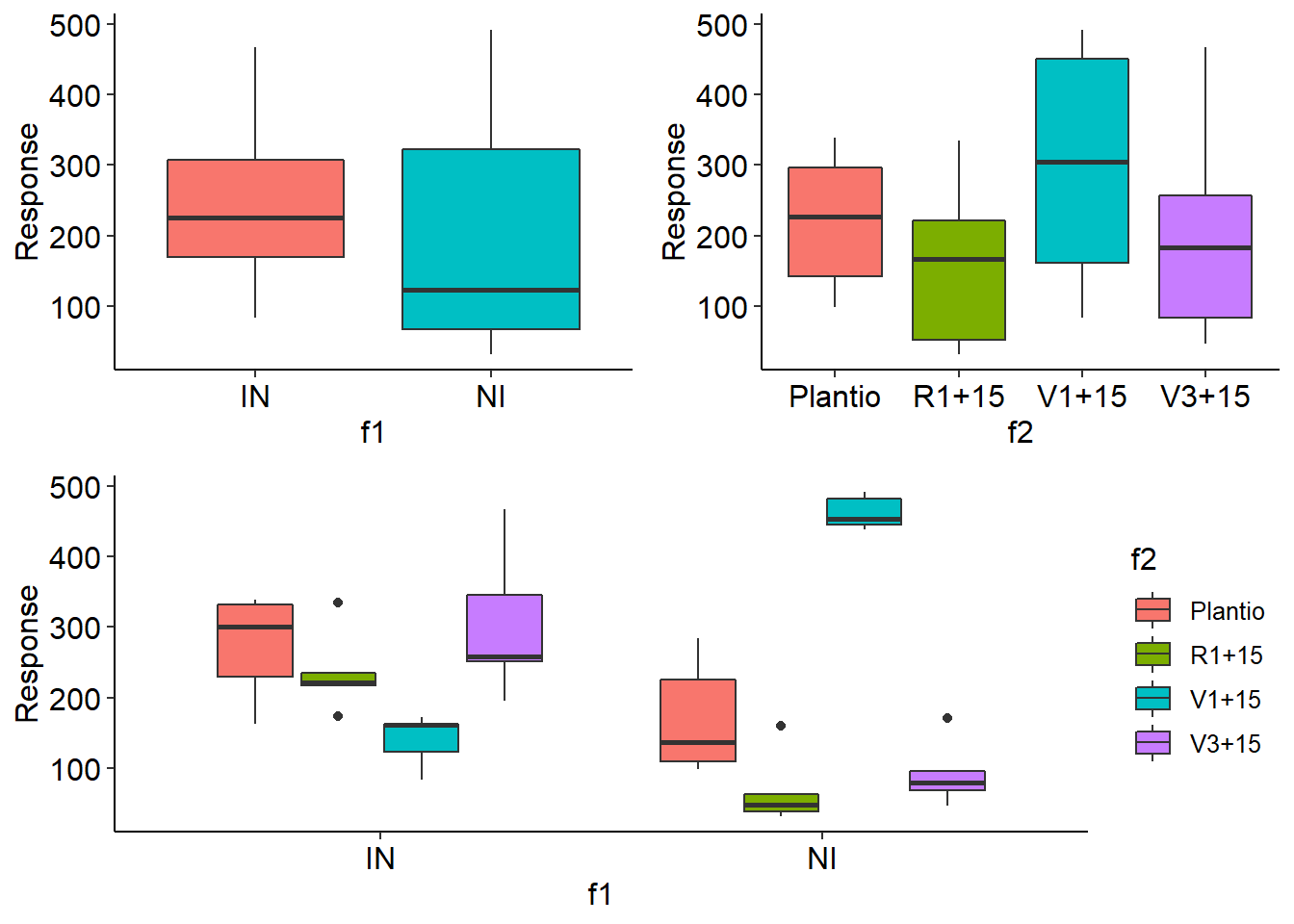
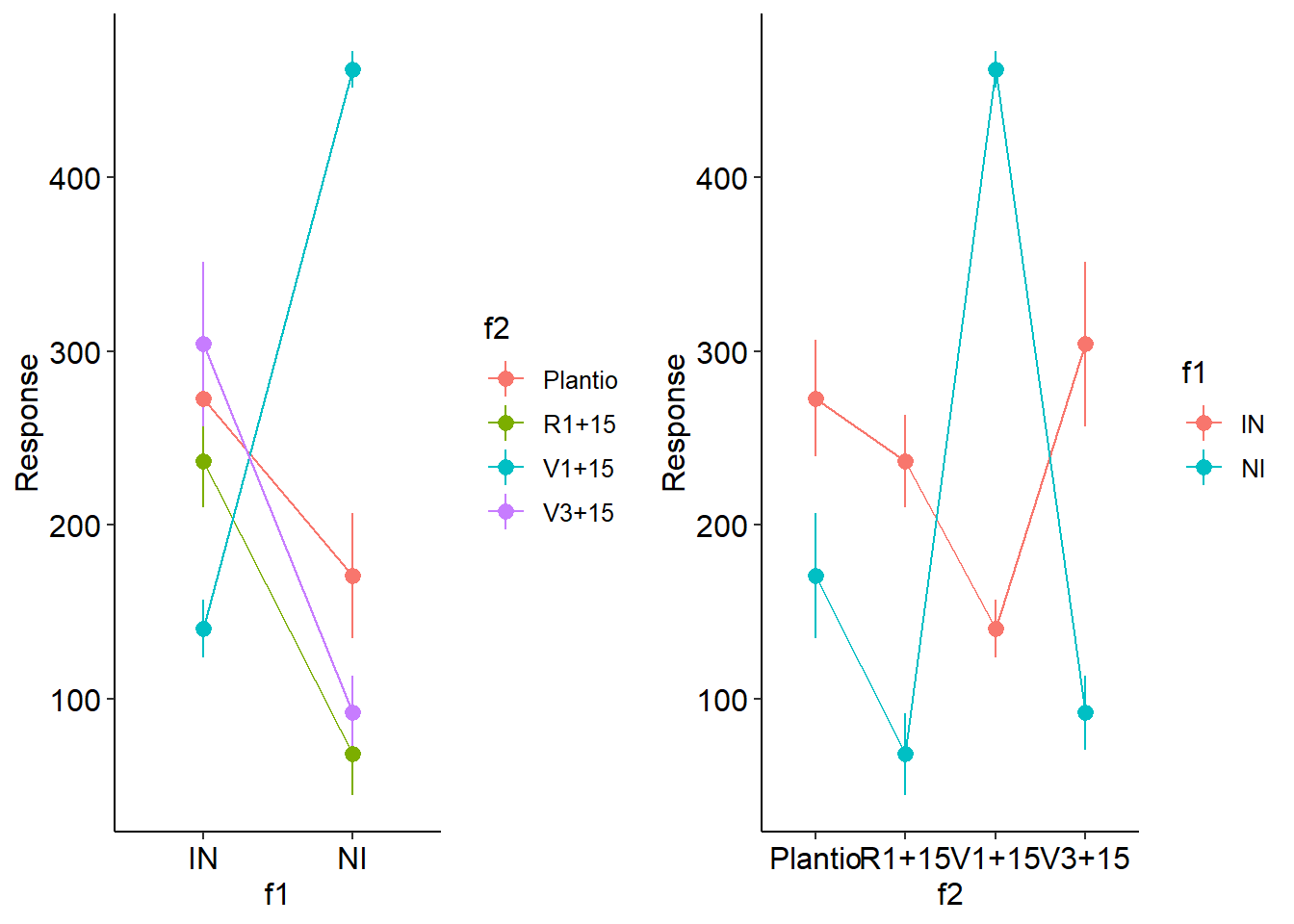
desc2fat(f1,f2,resp)
##
## -----------------------------------------------------------------
## general description
## -----------------------------------------------------------------
## Media Mediana Minimo Maximo Variancia Desvio CV
## [1,] 218.35 185 32 492 18474.95 135.9226 62.24987
##
## -----------------------------------------------------------------
## Interaction
## -----------------------------------------------------------------
## $Mean
## Plantio R1+15 V1+15 V3+15
## IN 272.8 236.6 140.4 304
## NI 170.6 68.2 462.2 92
##
## $Median
## Plantio R1+15 V1+15 V3+15
## IN 300 222 161 258
## NI 136 48 453 79
##
## $Min
## Plantio R1+15 V1+15 V3+15
## IN 163 174 83 196
## NI 98 32 438 46
##
## $Max
## Plantio R1+15 V1+15 V3+15
## IN 339 335 172 468
## NI 284 160 492 171
##
## $Variance
## Plantio R1+15 V1+15 V3+15
## IN 5628.7 3550.3 1382.8 11286
## NI 6489.8 2771.2 553.2 2266
##
## $SD
## Plantio R1+15 V1+15 V3+15
## IN 75.02466 59.58439 37.18602 106.23559
## NI 80.55929 52.64219 23.52020 47.60252
##
## $`CV(%)`
## Plantio R1+15 V1+15 V3+15
## IN 27.50171 25.18360 26.48577 34.94592
## NI 47.22116 77.18796 5.08875 51.74187
##
##
## -----------------------------------------------------------------
## f1
## -----------------------------------------------------------------
## Mean Median Min Max Variance SD CV(%)
## IN 238.45 226 83 468 8571.629 92.58309 38.82705
## NI 198.25 123 32 492 28500.092 168.81970 85.15496
##
## -----------------------------------------------------------------
## f2
## -----------------------------------------------------------------
## Mean Median Min Max Variance SD CV(%)
## Plantio 221.7 227.5 98 339 8287.344 91.03485 41.06218
## R1+15 152.4 167.0 32 335 10686.933 103.37762 67.83309
## V1+15 301.3 305.0 83 492 29625.789 172.12144 57.12627
## V3+15 198.0 183.5 46 468 18507.556 136.04248 68.70832
##
## -----------------------------------------------------------------
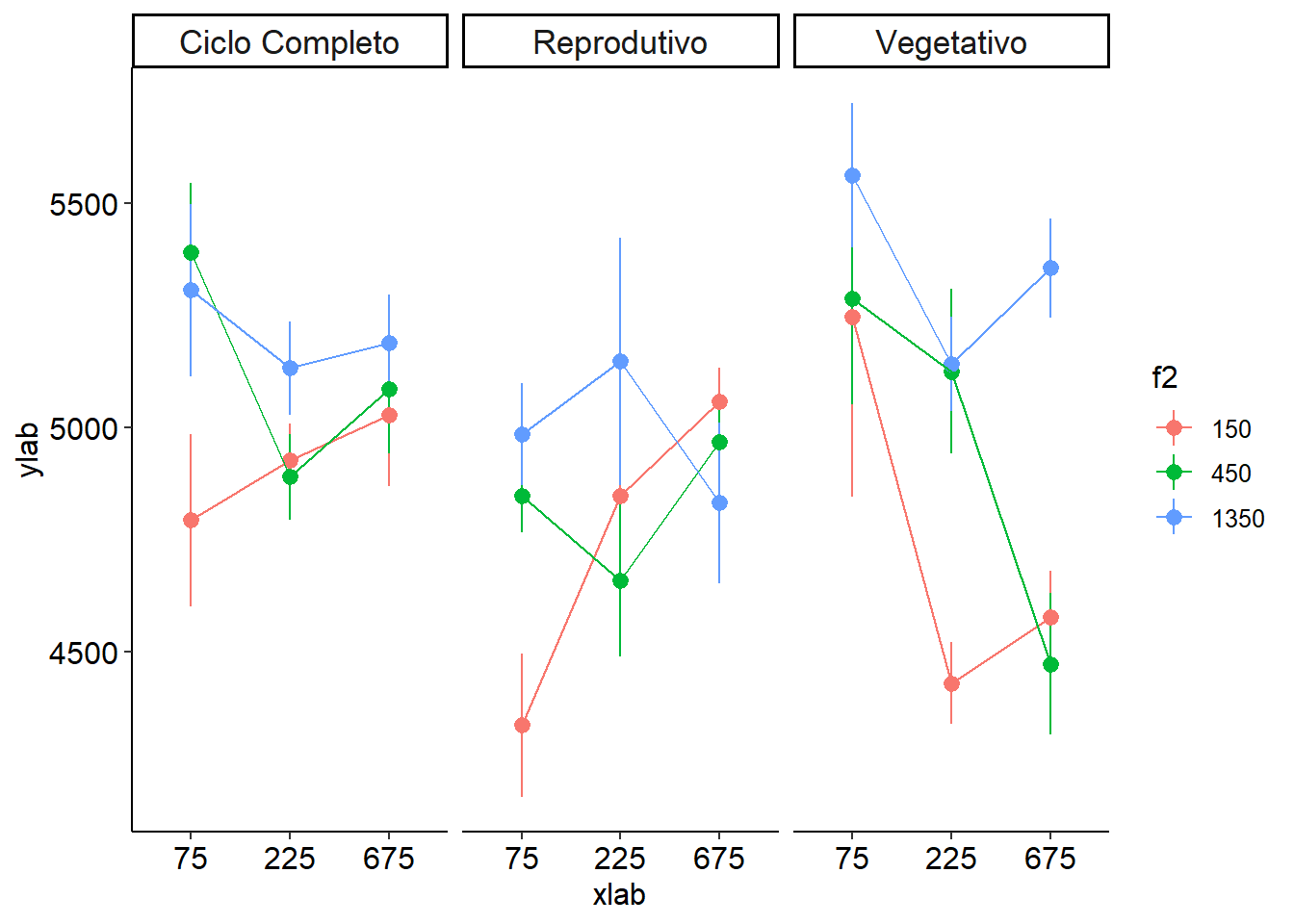
Três fatores
data(enxofre)
attach(enxofre)
## The following objects are masked from cloro:
##
## bloco, f1, f2, resp
desc3fat(f1, f2, f3, resp)
dispvar
data("pomegranate")
dispvar(pomegranate[,-1])
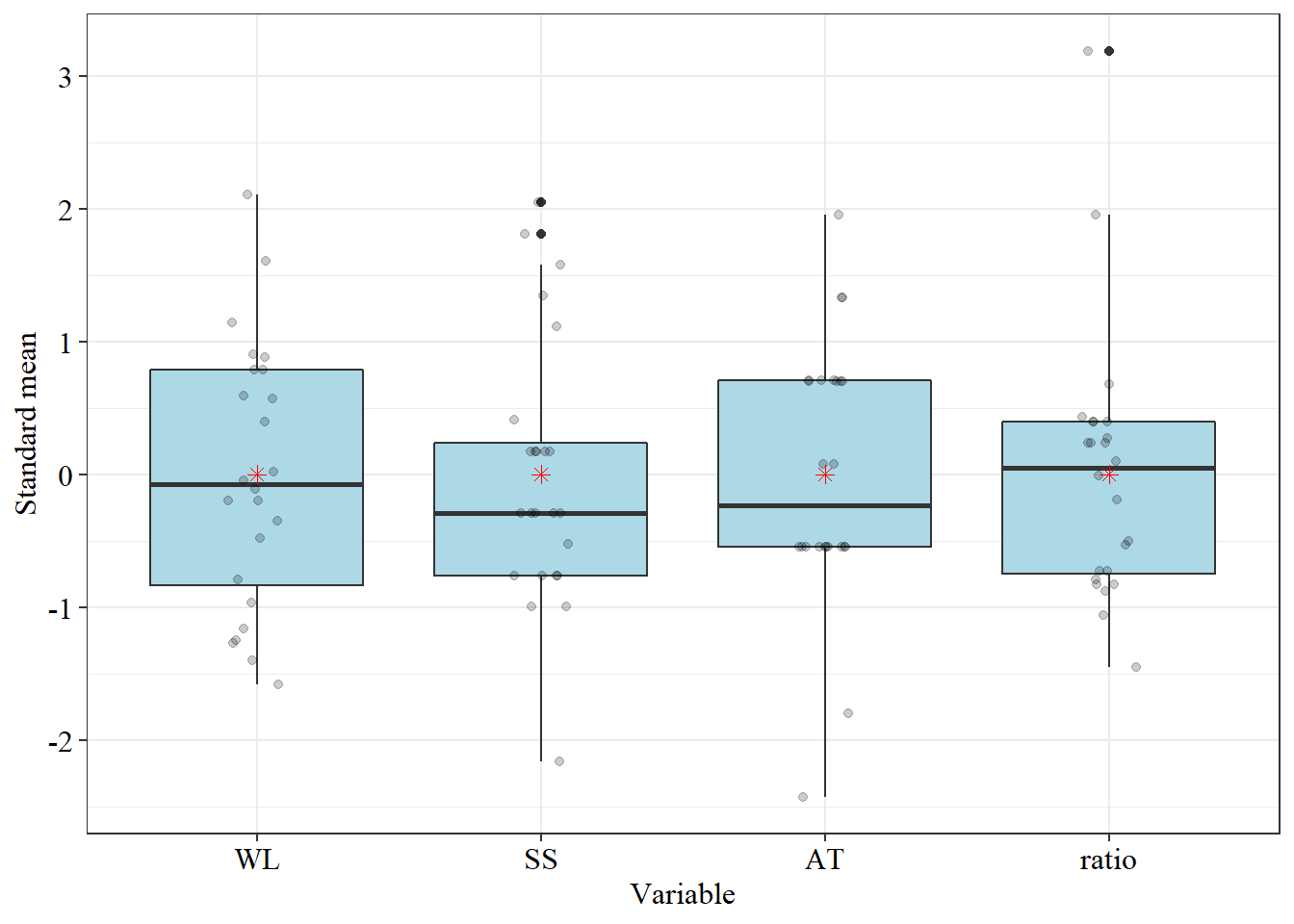
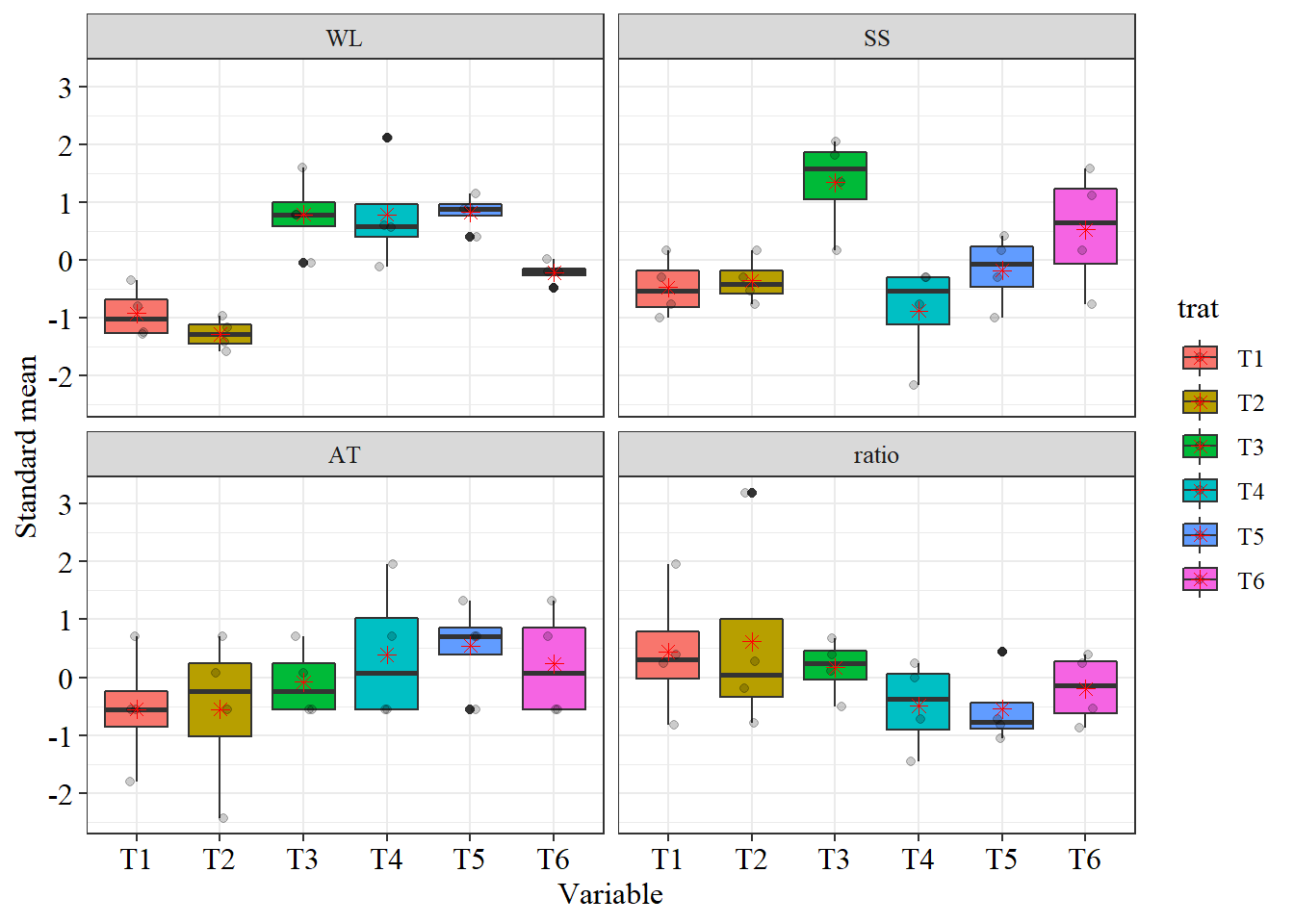
trat=pomegranate$trat
dispvar(pomegranate[,-1], trat)
tabledesc
data(pomegranate)
tabledesc(pomegranate)
## WL SS AT ratio
## T1 1.8425 14.225 0.900 16.23341
## T2 1.6775 14.275 0.900 16.73232
## T3 2.6175 15.000 0.975 15.48409
## T4 2.6200 14.050 1.050 13.66667
## T5 2.6375 14.350 1.075 13.50821
## T6 2.1625 14.650 1.025 14.47664





