Analysis: Randomized block design by glm
DBC.glm.RdStatistical analysis of experiments conducted in a randomized block design using a generalized linear model. It performs the deviance analysis and the effect is tested by a chi-square test. Multiple comparisons are adjusted by Tukey.
DBC.glm(
trat,
block,
response,
glm.family = "binomial",
quali = TRUE,
alpha.f = 0.05,
alpha.t = 0.05,
geom = "bar",
theme = theme_classic(),
sup = NA,
ylab = "Response",
xlab = "",
fill = "lightblue",
angle = 0,
family = "sans",
textsize = 12,
labelsize = 5,
dec = 3,
addmean = TRUE,
errorbar = TRUE,
posi = "top",
point = "mean_sd",
angle.label = 0
)Arguments
- trat
Numerical or complex vector with treatments
- block
Numerical or complex vector with blocks
- response
Numerical vector containing the response of the experiment. Use cbind(resp, n-resp) for binomial or quasibinomial family.
- glm.family
distribution family considered (default is binomial)
- quali
Defines whether the factor is quantitative or qualitative (default is qualitative)
- alpha.f
Level of significance of the F test (default is 0.05)
- alpha.t
Significance level of the multiple comparison test (default is 0.05)
- geom
Graph type (columns, boxes or segments)
- theme
ggplot2 theme (default is theme_classic())
- sup
Number of units above the standard deviation or average bar on the graph
- ylab
Variable response name (Accepts the expression() function)
- xlab
Treatments name (Accepts the expression() function)
- fill
Defines chart color (to generate different colors for different treatments, define fill = "trat")
- angle
x-axis scale text rotation
- family
Font family
- textsize
Font size
- labelsize
Label size
- dec
Number of cells
- addmean
Plot the average value on the graph (default is TRUE)
- errorbar
Plot the standard deviation bar on the graph (In the case of a segment and column graph) - default is TRUE
- posi
Legend position
- point
Defines whether to plot mean ("mean"), mean with standard deviation ("mean_sd" - default) or mean with standard error (default - "mean_se").
- angle.label
label angle
Examples
data("aristolochia")
attach(aristolochia)
#> The following object is masked from cloro:
#>
#> resp
#> The following object is masked from passiflora:
#>
#> trat
# Assuming the same aristolochia data set, but considering randomized blocks
bloco=rep(paste("B",1:16),5)
resp=resp/2
DBC.glm(trat,bloco, cbind(resp,50-resp), glm.family="binomial")
#>
#>
#> -----------------------------------------------------------------
#> Analysis of deviance
#> -----------------------------------------------------------------
#>
#> Null deviance 2159.834
#> Df Null deviance 79.000
#> -----
#> Treatment effects
#> Residual deviance 346.688
#> Df residual deviance 75.000
#> p-value(Chisq) 0.000
#> -----
#> Block effects
#> Residual deviance 247.739
#> Df residual deviance 60.000
#> p-value(Chisq) 0.000
#> -----
#> AIC 510.227
#>
#>
#> As the calculated p-value, it is less than the 5% significance level.The hypothesis H0 of equality of means is rejected. Therefore, at least two treatments differ
#>
#>
#> -----------------------------------------------------------------
#> Multiple Comparison Test
#> -----------------------------------------------------------------
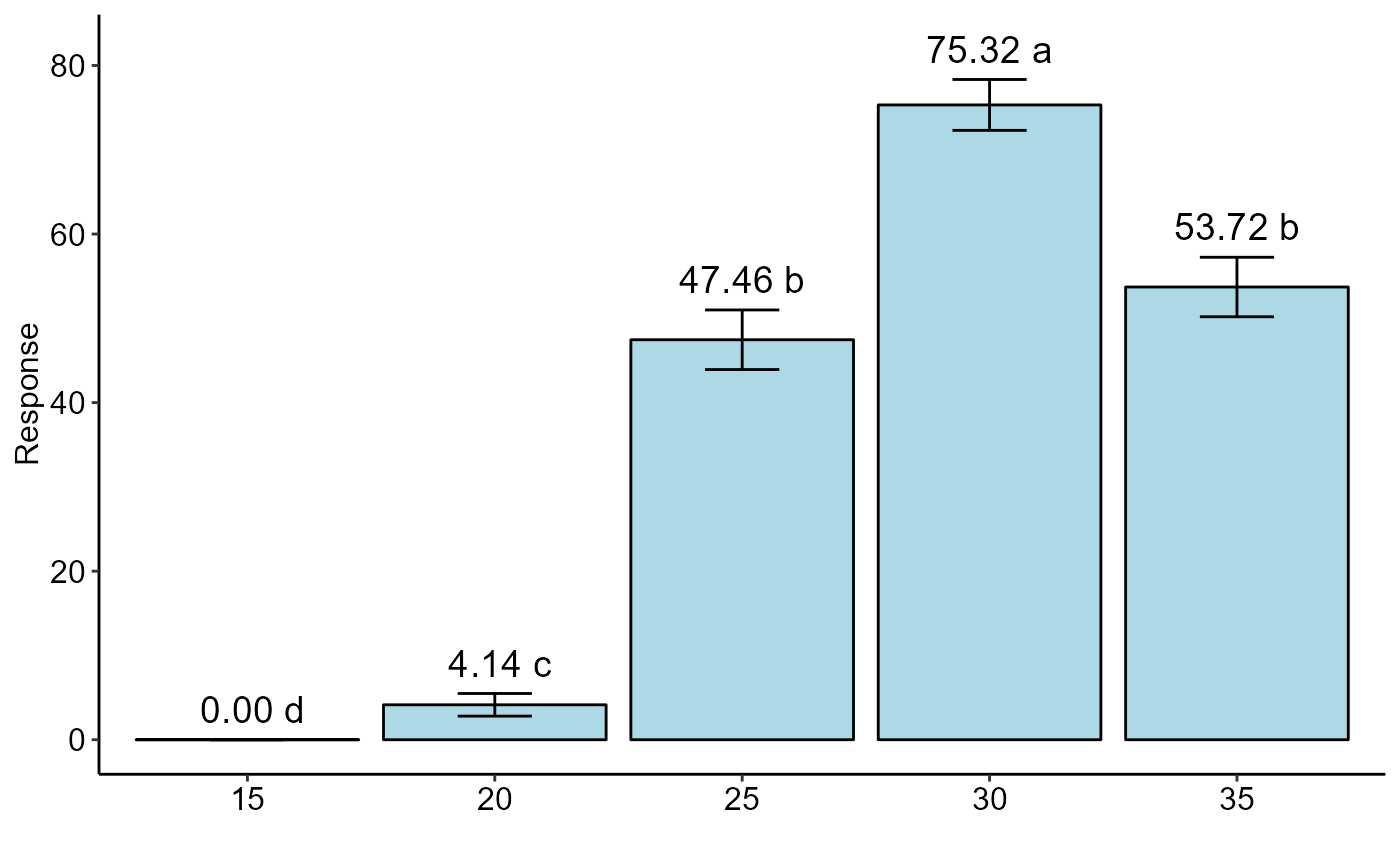
#> trat prob SE asymp.LCL asymp.UCL .group
#> 15 15 0.00 0.00 0.00 0.00 d
#> 20 20 0.04 0.01 0.03 0.05 c
#> 25 25 0.47 0.02 0.44 0.51 b
#> 30 30 0.75 0.02 0.72 0.78 a
#> 35 35 0.54 0.02 0.50 0.57 b