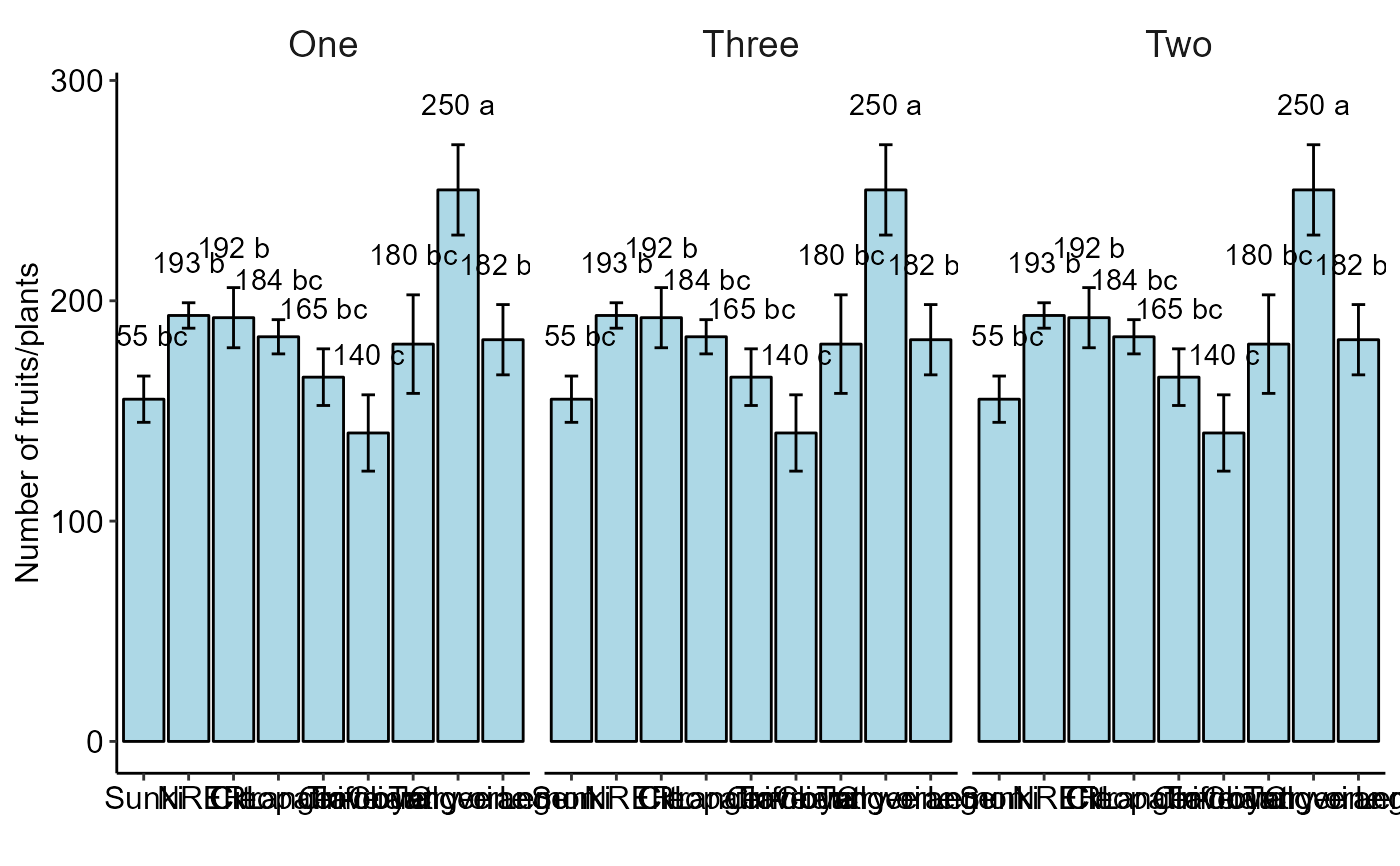
Graph: Group DIC, DBC and DQL functions column charts
bargraph_onefactor.RdGroups two or more column charts exported from DIC, DBC or DQL function
bargraph_onefactor(
analysis,
labels = NULL,
ocult.facet = FALSE,
ocult.box = FALSE,
facet.size = 14,
ylab = NULL,
width.bar = 0.3,
sup = NULL
)Arguments
- analysis
List with DIC, DBC or DQL object
- labels
Vector with the name of the facets
- ocult.facet
Hide facets
- ocult.box
Hide box
- facet.size
Font size facets
- ylab
Y-axis name
- width.bar
Width bar
- sup
Number of units above the standard deviation or average bar on the graph
Value
Returns a column chart grouped by facets
Examples
library(AgroR)
data("laranja")
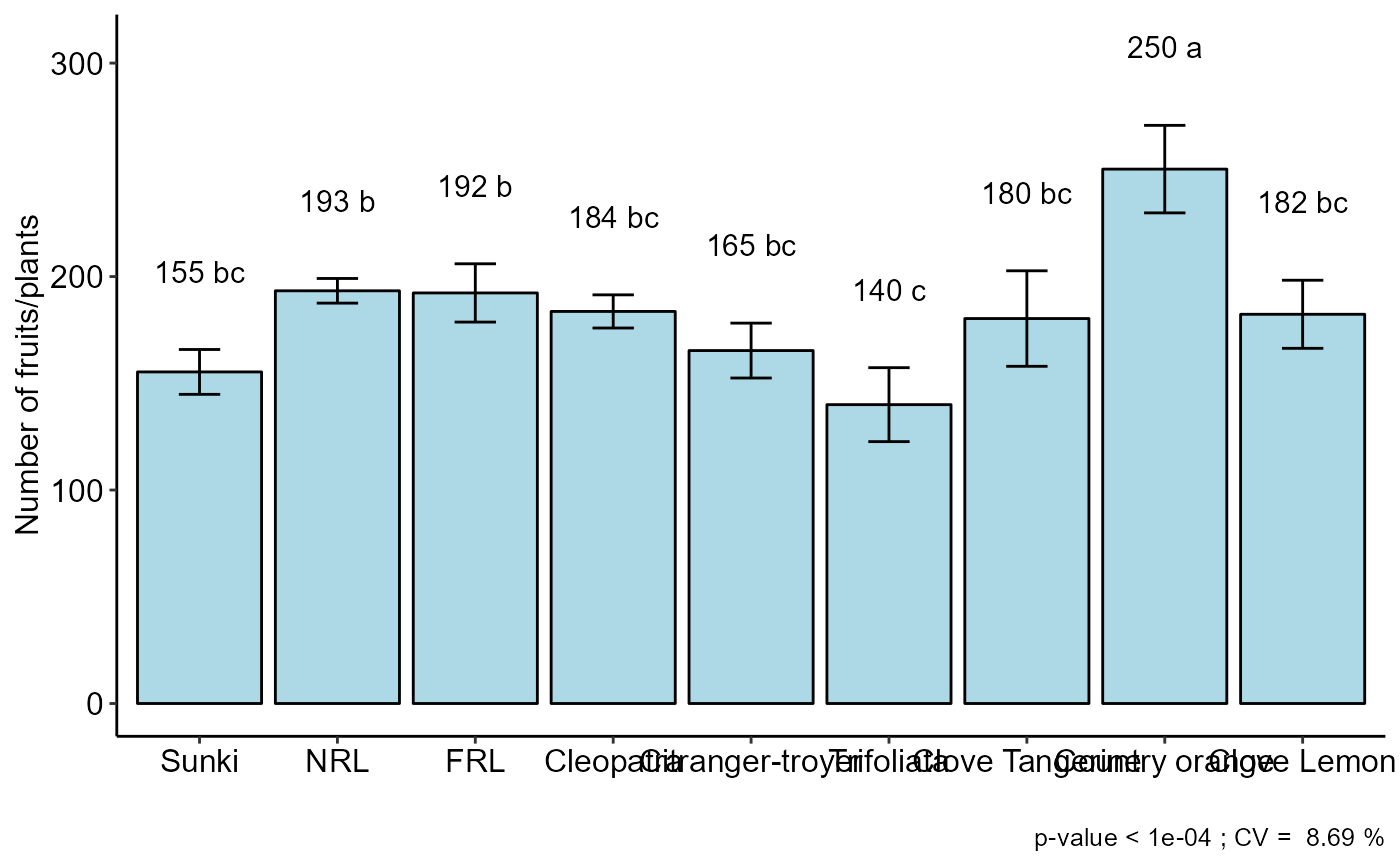
a=with(laranja, DBC(trat, bloco, resp, ylab = "Number of fruits/plants"))
#>
#> -----------------------------------------------------------------
#> Normality of errors
#> -----------------------------------------------------------------
#> Method Statistic p.value
#> Shapiro-Wilk normality test(W) 0.9475889 0.187264
#>
#> As the calculated p-value is greater than the 5% significance level, hypothesis H0 is not rejected. Therefore, errors can be considered normal
#>
#> -----------------------------------------------------------------
#> Homogeneity of Variances
#> -----------------------------------------------------------------
#> Method Statistic p.value
#> Bartlett test(Bartlett's K-squared) 4.036888 0.85378
#>
#> As the calculated p-value is greater than the 5% significance level, hypothesis H0 is not rejected. Therefore, the variances can be considered homogeneous
#>
#> -----------------------------------------------------------------
#> Independence from errors
#> -----------------------------------------------------------------
#> Method Statistic p.value
#> Durbin-Watson test(DW) 2.324604 0.2484349
#>
#> As the calculated p-value is greater than the 5% significance level, hypothesis H0 is not rejected. Therefore, errors can be considered independent
#>
#> -----------------------------------------------------------------
#> Additional Information
#> -----------------------------------------------------------------
#>
#> CV (%) = 8.69
#> MStrat/MST = 0.91
#> Mean = 182.5556
#> Median = 183
#> Possible outliers = No discrepant point
#>
#> -----------------------------------------------------------------
#> Analysis of Variance
#> -----------------------------------------------------------------
#> Df Sum Sq Mean.Sq F value Pr(F)
#> trat 8 22981.33333 2872.66667 11.41142069 2.636524e-05
#> bloco 2 33.55556 16.77778 0.06664828 9.357825e-01
#> Residuals 16 4027.77778 251.73611
#>
#> As the calculated p-value, it is less than the 5% significance level. The hypothesis H0 of equality of means is rejected. Therefore, at least two treatments differ
 #>
#> -----------------------------------------------------------------
#> Multiple Comparison Test: Tukey HSD
#> -----------------------------------------------------------------
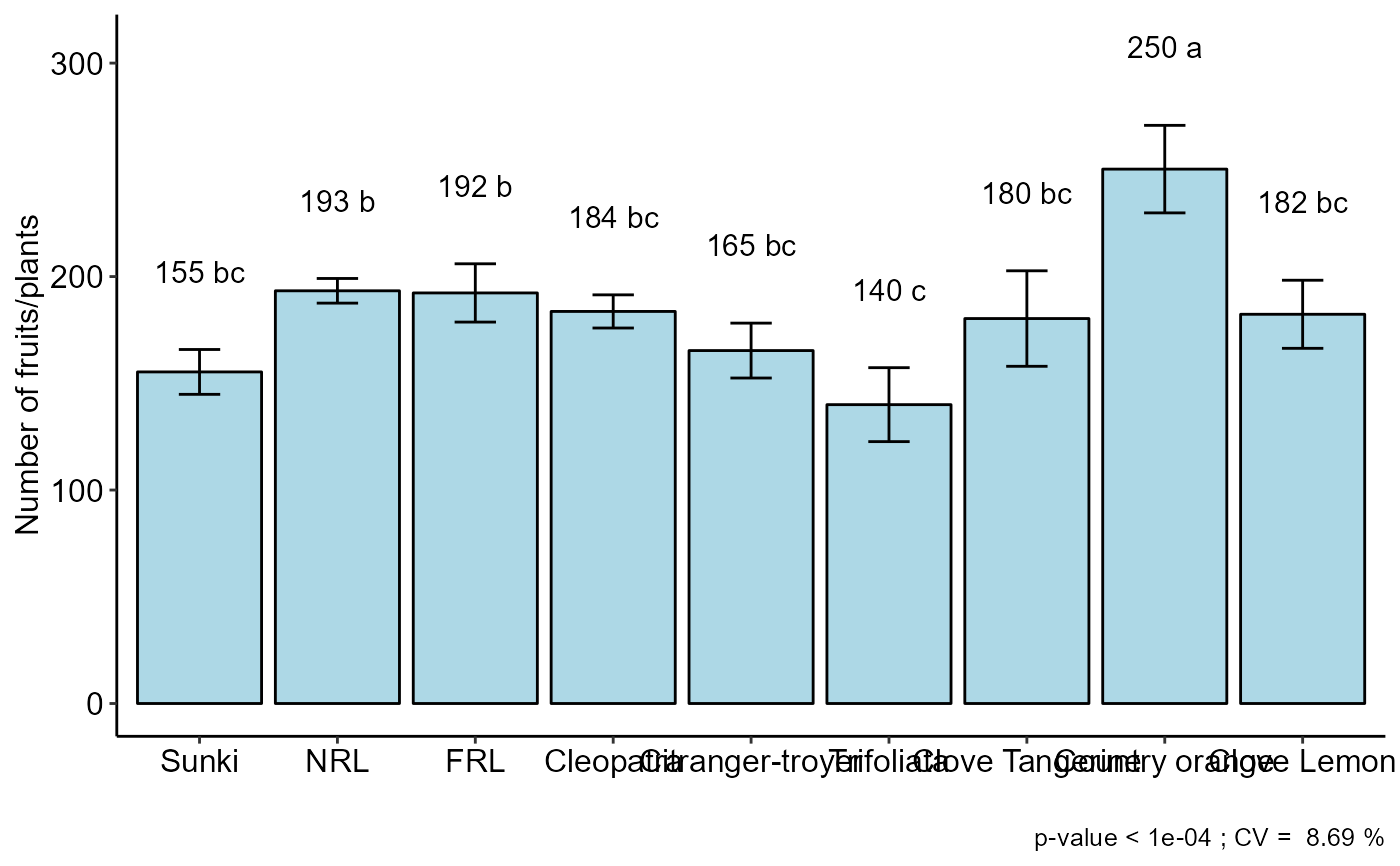
#> resp groups
#> Country orange 250.3333 a
#> NRL 193.3333 b
#> FRL 192.3333 b
#> Cleopatra 183.6667 bc
#> Clove Lemon 182.3333 bc
#> Clove Tangerine 180.3333 bc
#> Citranger-troyer 165.3333 bc
#> Sunki 155.3333 bc
#> Trifoliata 140.0000 c
#>
#>
#> -----------------------------------------------------------------
#> Multiple Comparison Test: Tukey HSD
#> -----------------------------------------------------------------
#> resp groups
#> Country orange 250.3333 a
#> NRL 193.3333 b
#> FRL 192.3333 b
#> Cleopatra 183.6667 bc
#> Clove Lemon 182.3333 bc
#> Clove Tangerine 180.3333 bc
#> Citranger-troyer 165.3333 bc
#> Sunki 155.3333 bc
#> Trifoliata 140.0000 c
#>
 b=with(laranja, DBC(trat, bloco, resp, ylab = "Number of fruits/plants"))
#>
#> -----------------------------------------------------------------
#> Normality of errors
#> -----------------------------------------------------------------
#> Method Statistic p.value
#> Shapiro-Wilk normality test(W) 0.9475889 0.187264
#>
#> As the calculated p-value is greater than the 5% significance level, hypothesis H0 is not rejected. Therefore, errors can be considered normal
#>
#> -----------------------------------------------------------------
#> Homogeneity of Variances
#> -----------------------------------------------------------------
#> Method Statistic p.value
#> Bartlett test(Bartlett's K-squared) 4.036888 0.85378
#>
#> As the calculated p-value is greater than the 5% significance level, hypothesis H0 is not rejected. Therefore, the variances can be considered homogeneous
#>
#> -----------------------------------------------------------------
#> Independence from errors
#> -----------------------------------------------------------------
#> Method Statistic p.value
#> Durbin-Watson test(DW) 2.324604 0.2484349
#>
#> As the calculated p-value is greater than the 5% significance level, hypothesis H0 is not rejected. Therefore, errors can be considered independent
#>
#> -----------------------------------------------------------------
#> Additional Information
#> -----------------------------------------------------------------
#>
#> CV (%) = 8.69
#> MStrat/MST = 0.91
#> Mean = 182.5556
#> Median = 183
#> Possible outliers = No discrepant point
#>
#> -----------------------------------------------------------------
#> Analysis of Variance
#> -----------------------------------------------------------------
#> Df Sum Sq Mean.Sq F value Pr(F)
#> trat 8 22981.33333 2872.66667 11.41142069 2.636524e-05
#> bloco 2 33.55556 16.77778 0.06664828 9.357825e-01
#> Residuals 16 4027.77778 251.73611
#>
#> As the calculated p-value, it is less than the 5% significance level. The hypothesis H0 of equality of means is rejected. Therefore, at least two treatments differ
b=with(laranja, DBC(trat, bloco, resp, ylab = "Number of fruits/plants"))
#>
#> -----------------------------------------------------------------
#> Normality of errors
#> -----------------------------------------------------------------
#> Method Statistic p.value
#> Shapiro-Wilk normality test(W) 0.9475889 0.187264
#>
#> As the calculated p-value is greater than the 5% significance level, hypothesis H0 is not rejected. Therefore, errors can be considered normal
#>
#> -----------------------------------------------------------------
#> Homogeneity of Variances
#> -----------------------------------------------------------------
#> Method Statistic p.value
#> Bartlett test(Bartlett's K-squared) 4.036888 0.85378
#>
#> As the calculated p-value is greater than the 5% significance level, hypothesis H0 is not rejected. Therefore, the variances can be considered homogeneous
#>
#> -----------------------------------------------------------------
#> Independence from errors
#> -----------------------------------------------------------------
#> Method Statistic p.value
#> Durbin-Watson test(DW) 2.324604 0.2484349
#>
#> As the calculated p-value is greater than the 5% significance level, hypothesis H0 is not rejected. Therefore, errors can be considered independent
#>
#> -----------------------------------------------------------------
#> Additional Information
#> -----------------------------------------------------------------
#>
#> CV (%) = 8.69
#> MStrat/MST = 0.91
#> Mean = 182.5556
#> Median = 183
#> Possible outliers = No discrepant point
#>
#> -----------------------------------------------------------------
#> Analysis of Variance
#> -----------------------------------------------------------------
#> Df Sum Sq Mean.Sq F value Pr(F)
#> trat 8 22981.33333 2872.66667 11.41142069 2.636524e-05
#> bloco 2 33.55556 16.77778 0.06664828 9.357825e-01
#> Residuals 16 4027.77778 251.73611
#>
#> As the calculated p-value, it is less than the 5% significance level. The hypothesis H0 of equality of means is rejected. Therefore, at least two treatments differ
 #>
#> -----------------------------------------------------------------
#> Multiple Comparison Test: Tukey HSD
#> -----------------------------------------------------------------
#> resp groups
#> Country orange 250.3333 a
#> NRL 193.3333 b
#> FRL 192.3333 b
#> Cleopatra 183.6667 bc
#> Clove Lemon 182.3333 bc
#> Clove Tangerine 180.3333 bc
#> Citranger-troyer 165.3333 bc
#> Sunki 155.3333 bc
#> Trifoliata 140.0000 c
#>
#>
#> -----------------------------------------------------------------
#> Multiple Comparison Test: Tukey HSD
#> -----------------------------------------------------------------
#> resp groups
#> Country orange 250.3333 a
#> NRL 193.3333 b
#> FRL 192.3333 b
#> Cleopatra 183.6667 bc
#> Clove Lemon 182.3333 bc
#> Clove Tangerine 180.3333 bc
#> Citranger-troyer 165.3333 bc
#> Sunki 155.3333 bc
#> Trifoliata 140.0000 c
#>
 c=with(laranja, DBC(trat, bloco, resp, ylab = "Number of fruits/plants"))
#>
#> -----------------------------------------------------------------
#> Normality of errors
#> -----------------------------------------------------------------
#> Method Statistic p.value
#> Shapiro-Wilk normality test(W) 0.9475889 0.187264
#>
#> As the calculated p-value is greater than the 5% significance level, hypothesis H0 is not rejected. Therefore, errors can be considered normal
#>
#> -----------------------------------------------------------------
#> Homogeneity of Variances
#> -----------------------------------------------------------------
#> Method Statistic p.value
#> Bartlett test(Bartlett's K-squared) 4.036888 0.85378
#>
#> As the calculated p-value is greater than the 5% significance level, hypothesis H0 is not rejected. Therefore, the variances can be considered homogeneous
#>
#> -----------------------------------------------------------------
#> Independence from errors
#> -----------------------------------------------------------------
#> Method Statistic p.value
#> Durbin-Watson test(DW) 2.324604 0.2484349
#>
#> As the calculated p-value is greater than the 5% significance level, hypothesis H0 is not rejected. Therefore, errors can be considered independent
#>
#> -----------------------------------------------------------------
#> Additional Information
#> -----------------------------------------------------------------
#>
#> CV (%) = 8.69
#> MStrat/MST = 0.91
#> Mean = 182.5556
#> Median = 183
#> Possible outliers = No discrepant point
#>
#> -----------------------------------------------------------------
#> Analysis of Variance
#> -----------------------------------------------------------------
#> Df Sum Sq Mean.Sq F value Pr(F)
#> trat 8 22981.33333 2872.66667 11.41142069 2.636524e-05
#> bloco 2 33.55556 16.77778 0.06664828 9.357825e-01
#> Residuals 16 4027.77778 251.73611
#>
#> As the calculated p-value, it is less than the 5% significance level. The hypothesis H0 of equality of means is rejected. Therefore, at least two treatments differ
c=with(laranja, DBC(trat, bloco, resp, ylab = "Number of fruits/plants"))
#>
#> -----------------------------------------------------------------
#> Normality of errors
#> -----------------------------------------------------------------
#> Method Statistic p.value
#> Shapiro-Wilk normality test(W) 0.9475889 0.187264
#>
#> As the calculated p-value is greater than the 5% significance level, hypothesis H0 is not rejected. Therefore, errors can be considered normal
#>
#> -----------------------------------------------------------------
#> Homogeneity of Variances
#> -----------------------------------------------------------------
#> Method Statistic p.value
#> Bartlett test(Bartlett's K-squared) 4.036888 0.85378
#>
#> As the calculated p-value is greater than the 5% significance level, hypothesis H0 is not rejected. Therefore, the variances can be considered homogeneous
#>
#> -----------------------------------------------------------------
#> Independence from errors
#> -----------------------------------------------------------------
#> Method Statistic p.value
#> Durbin-Watson test(DW) 2.324604 0.2484349
#>
#> As the calculated p-value is greater than the 5% significance level, hypothesis H0 is not rejected. Therefore, errors can be considered independent
#>
#> -----------------------------------------------------------------
#> Additional Information
#> -----------------------------------------------------------------
#>
#> CV (%) = 8.69
#> MStrat/MST = 0.91
#> Mean = 182.5556
#> Median = 183
#> Possible outliers = No discrepant point
#>
#> -----------------------------------------------------------------
#> Analysis of Variance
#> -----------------------------------------------------------------
#> Df Sum Sq Mean.Sq F value Pr(F)
#> trat 8 22981.33333 2872.66667 11.41142069 2.636524e-05
#> bloco 2 33.55556 16.77778 0.06664828 9.357825e-01
#> Residuals 16 4027.77778 251.73611
#>
#> As the calculated p-value, it is less than the 5% significance level. The hypothesis H0 of equality of means is rejected. Therefore, at least two treatments differ
 #>
#> -----------------------------------------------------------------
#> Multiple Comparison Test: Tukey HSD
#> -----------------------------------------------------------------
#> resp groups
#> Country orange 250.3333 a
#> NRL 193.3333 b
#> FRL 192.3333 b
#> Cleopatra 183.6667 bc
#> Clove Lemon 182.3333 bc
#> Clove Tangerine 180.3333 bc
#> Citranger-troyer 165.3333 bc
#> Sunki 155.3333 bc
#> Trifoliata 140.0000 c
#>
#>
#> -----------------------------------------------------------------
#> Multiple Comparison Test: Tukey HSD
#> -----------------------------------------------------------------
#> resp groups
#> Country orange 250.3333 a
#> NRL 193.3333 b
#> FRL 192.3333 b
#> Cleopatra 183.6667 bc
#> Clove Lemon 182.3333 bc
#> Clove Tangerine 180.3333 bc
#> Citranger-troyer 165.3333 bc
#> Sunki 155.3333 bc
#> Trifoliata 140.0000 c
#>
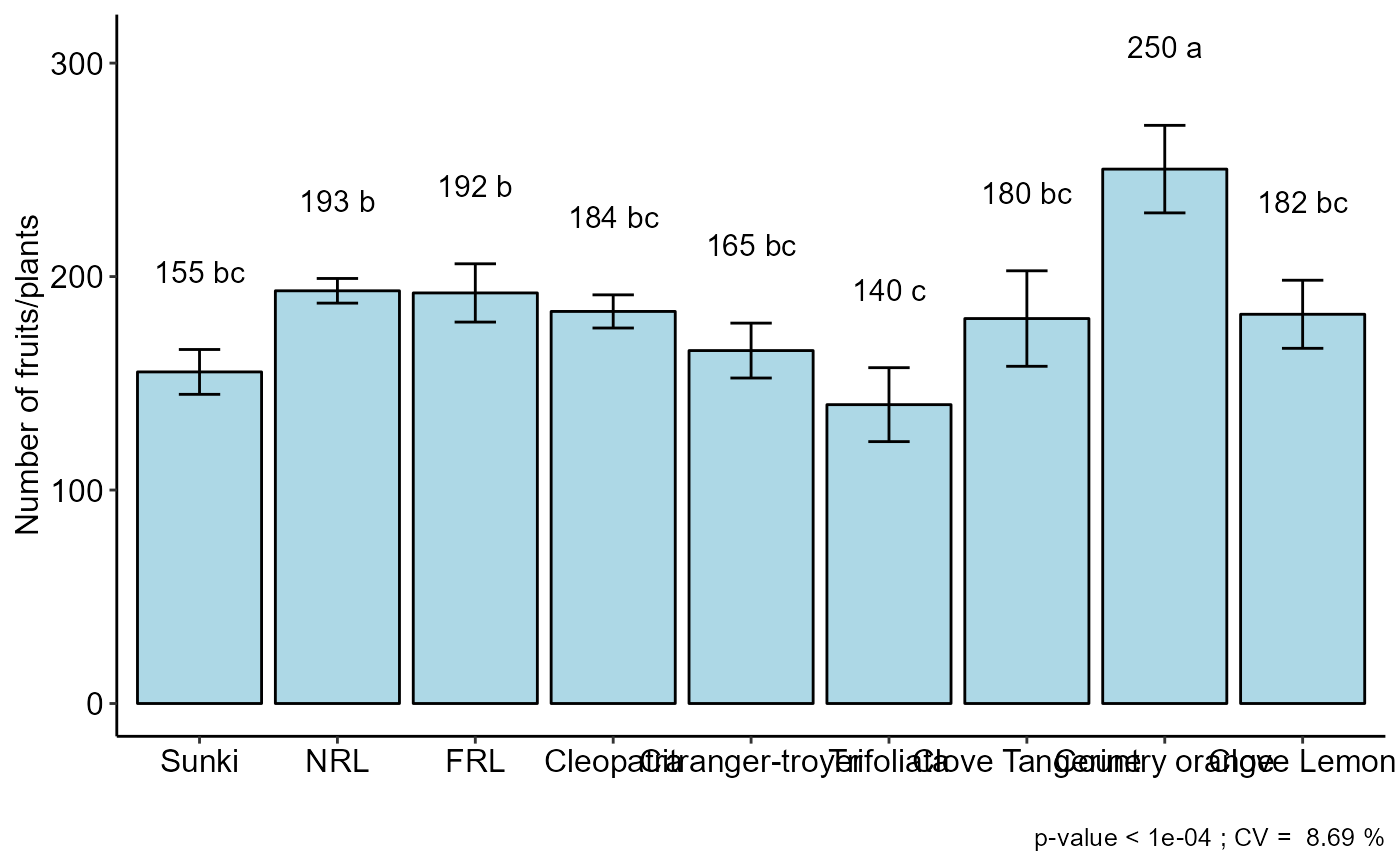 bargraph_onefactor(analysis = list(a,b,c), labels = c("One","Two","Three"),ocult.box = TRUE)
bargraph_onefactor(analysis = list(a,b,c), labels = c("One","Two","Three"),ocult.box = TRUE)